Time: 18th-23rd June 2019
Place: ETH, Zurich, Switzerland
Participation fee: 500 CHF (academia) / 1'000 CHF (industry)
- Anyone can apply - irrespective of affiliation, position, or location!
Applications accepted from: 2019-03-15
Application deadline: 2019-04-15 (23:59 CET)
Application answer: 2019-05-01
Application Closed
About the School
This one-week school provides a hands-on introduction to image processing and analysis, with an emphasis on biologically relevant examples
Is this school for you?
- Are you a life-science researcher with a pressing need to quantify your light-microscopy images?
- Are you uncertain about how to: Best calculate co-localisation, do deconvolution, automate the counting of cells, track objects over time, handle massive amounts of image data, record your image-analysis work-flows in a reproducible manner?
- If you answered yes to some of the above, then this school is for you!
Motivation
Digital images of high quality and quantity are now the norm in biomedical sciences. Ten to twenty years ago, when many current professors trained as students or post-docs, this was not yet the case as most microscopes were, at best, equipped with low-resolution digital cameras, celluloid-film (analogue) cameras, or no camera at all.
This rapid change is rarely reflected in the curricula of life-science university departments and they offer few, if any, courses in image processing and analysis. Understandably, courses in image-analysis (computer-vision) in the computer-science departments tend to have different aims, work on different image-data, and pre-suppose literacy in at least one programming language, rendering them all but irrelevant for the life-scientist working in the laboratory.
To bridge this gap, between what the life-scientist needs and what courses he/she is normally offered, we have created this school for image analysis. No former experience with programming is assumed, nor will much indeed be needed; only a strong desire to learn what can be done and how to do it is required.
What you will learn
You will learn the fundamentals of image analysis, including basic macro programming in ImageJ/Fiji as well as other software solutions.
In the first part of the week we will also cover the process of image-formation as it pertains to image analysis: Resolution, correct exposure, point-spread functions, detector noise, Shannon's sampling theorem, and aliasing. All with a clear focus on application in the lab.
In the second half of the week there will be a number of focused topics, building on what was learnt during the first days, possible topics are:
- Deep Learning for image restoration and segmentation
- Co-localisation, i.e., spatial correlation analysis and statistics
- SMLM (Single-Molecule Localisation Microscopy) data analysis
- De-convolution of microscopy images
- Tracking of particles and cells in time-lapse recordings
- Stitching and registration of stacks of large image data
- Hands-on training with image-analysis software solutions such as ilastik and CellProfiler
Structure of the week
You will be working actively with image-analysis software every day -- this is an interactive hands-on school, not a passive lecture series. Short introductions are followed by guided work-flows that we step through together. You can and should ask questions at any time throughout.
There will be a single invited lecture every day, alternating between scientists using image-analysis as an integral part of their biomedical research and researchers developing new image-analysis algorithms and software.
You will have the chance to work on your own data, that you brought from home, throughout the week, with support from the trainers.
In the second part of the week you will be working on image-analysis projects, in groups, with support from the trainers. On the last day you will present the results of your project-work.
Printed material as well as online material will be made available for all the modules.
Factoids
- We will accept 25 participants --- this number is kept low to facilitate effective tutoring.
- The school takes place in Zurich
- You will need to bring your own laptop (let us know if this is not possible for you)
- Participation is only possible for the entire week
- A small number or travel grants and fee-waivers are available, mainly for non-Swiss people
- PhD students can earn two ECTS points from the school
- Refreshments will be provided, meals are up to you
- Travel and accommodation arrangements are your own responsibility
- All teaching takes place in the ETH main building: Rämistrasse 101, 8092 Zurich, Switzerland
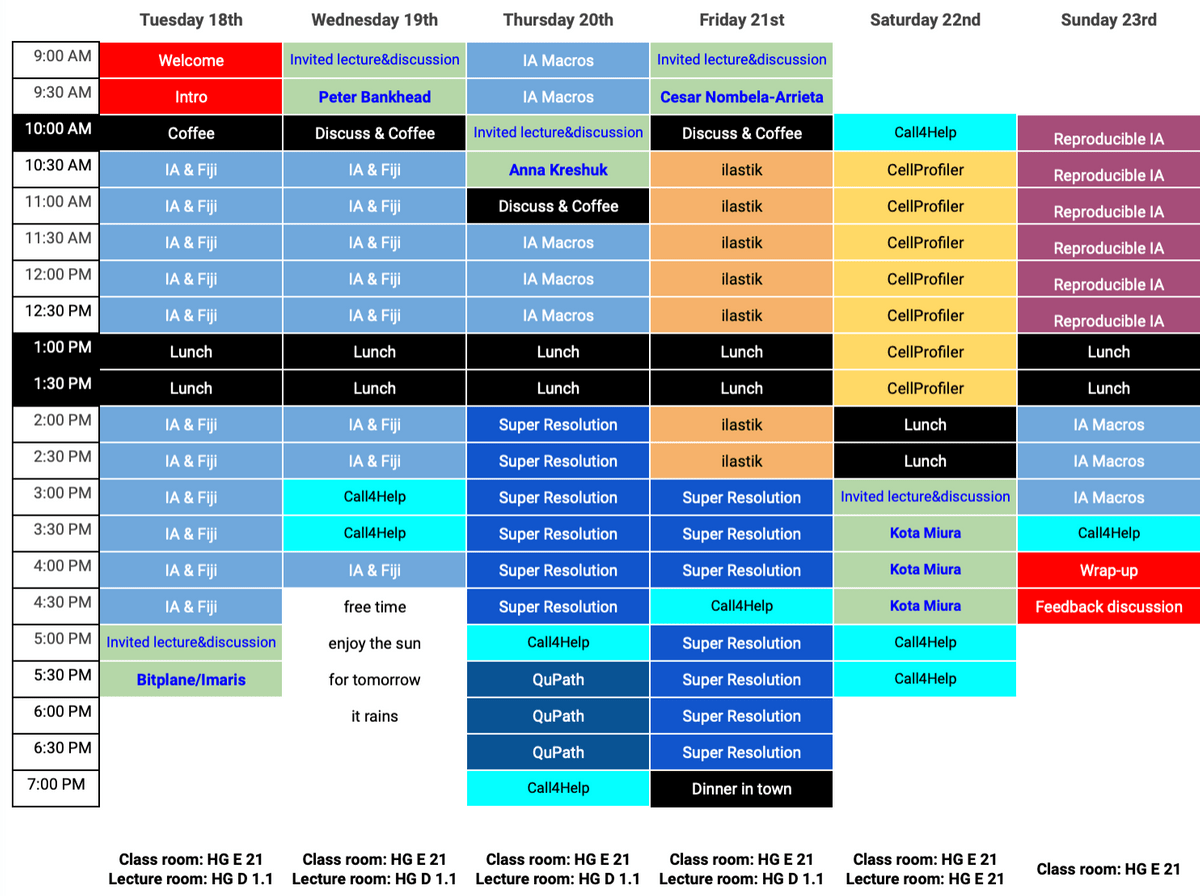
Program
Expect some changes as we adapt the pace dynamically

Full address: ETH Zürich; Main building; Rämistrasse 101; 8092 Zürich; Switzerland
Poster
Please help us promote the event!
Trainers
With backgrounds in biology, computer science, and physics and extensive teaching experience across the disciplines


2019 Speakers
Scientists using or developing image analysis methods in their research
My main research focus is bioimage analysis and digital pathology. I am especially interested in developing new and practical approaches to analyse whole slide scans of tissue, collaborating with pathologists and other researchers to apply these methods to answer important biomedical questions.
The Kreshuk group develops machine learning-based methods and tools for automatic segmentation, classification and analysis of biological images.
Our lab is interested in studying how the heterogeneous constituents of mammalian bone marrow (BM) tissues are structurally and functionally interconnected to work as a single finely-tuned, sophisticated and versatile functional unit.

Dr. Kota Miura
EMBL / University of Heidelberg, Heidelberg
Kota Miura had been working at the Centre for Molecular and Cellular Imaging, EMBL Heidelberg as Scientist and IT engineer since 2005, and since 2014, he is working as an associate professor of the European branch office of National Institute of Basic Biology (Japan) and as a senior image analyst at EMBL. B.L.A. (Liberal Arts, ICU, Tokyo), M.Sc (Physiology, Osaka), Ph.D. (Cell and Developmental Biology, Munich). A biologist now very much specialized in image analysis.
Organisers
Scientific Organisers
Course Organisers
University and ETH Zurich
Advisory Board
Zurich Image and Data Analysis School 2019
Managing Director
ETH Zurich
Scientific Center for Optical and Electron MicroscopyTechnical Director - Light microscopy & screening
ETH ZurichConnect With Us
Something unclear? Let us know!
© 2019